Object segmentations with FastSAM and OpenVINO#
This Jupyter notebook can be launched on-line, opening an interactive environment in a browser window. You can also make a local installation. Choose one of the following options:
The Fast Segment Anything Model
(FastSAM) is a
real-time CNN-based model that can segment any object within an image
based on various user prompts. Segment Anything task is designed to
make vision tasks easier by providing an efficient way to identify
objects in an image. FastSAM significantly reduces computational demands
while maintaining competitive performance, making it a practical choice
for a variety of vision tasks.
FastSAM is a model that aims to overcome the limitations of the Segment Anything Model (SAM), which is a Transformer model that requires significant computational resources. FastSAM tackles the segment anything task by dividing it into two consecutive stages: all-instance segmentation and prompt-guided selection.
In the first stage, YOLOv8-seg is used to produce segmentation masks for all instances in the image. In the second stage, FastSAM outputs the region-of-interest corresponding to the prompt.
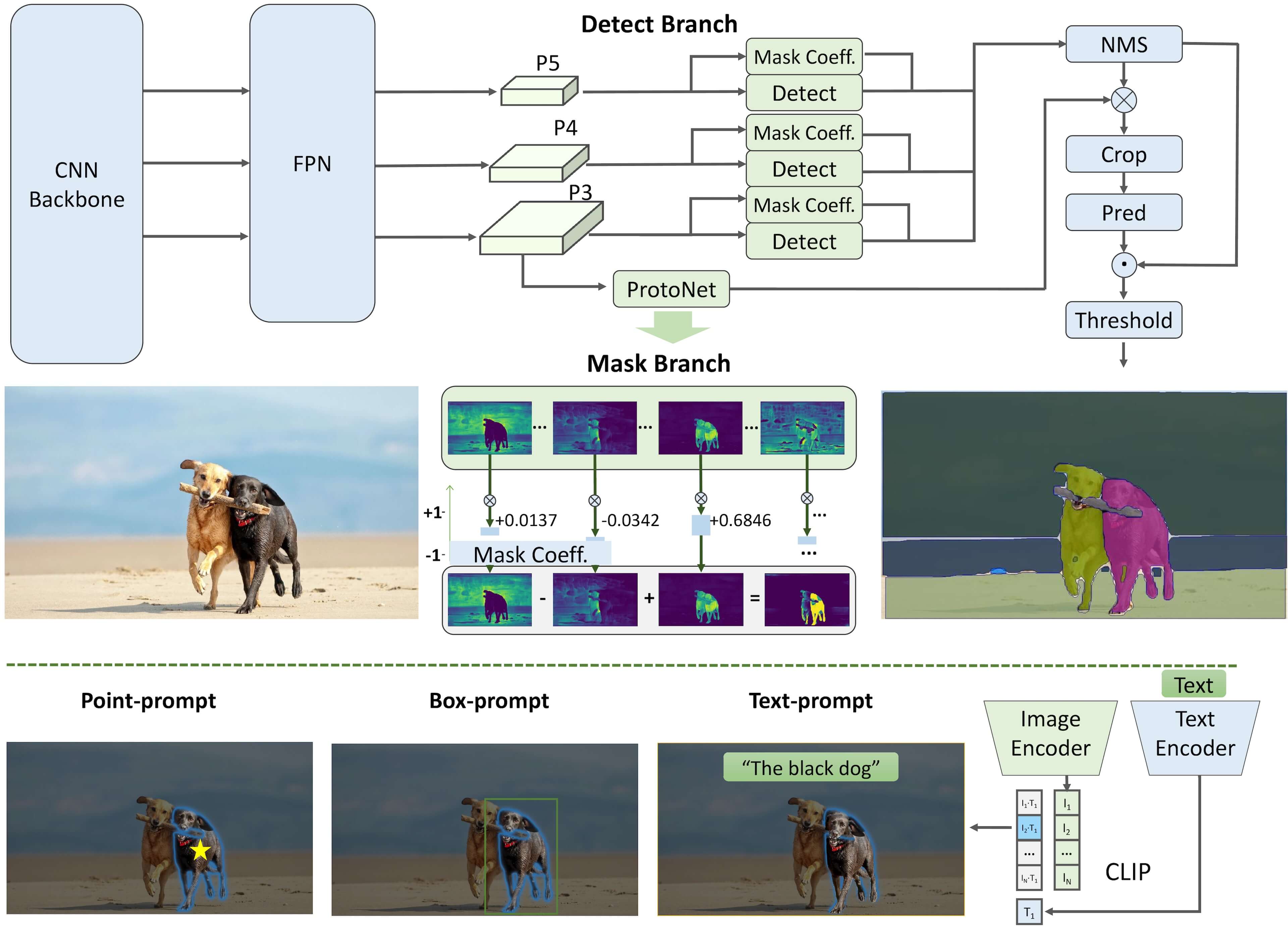
pipeline#
Table of contents:
Convert the model to OpenVINO Intermediate representation (IR) format
Optimize the model using NNCF Post-training Quantization API
Installation Instructions#
This is a self-contained example that relies solely on its own code.
We recommend running the notebook in a virtual environment. You only need a Jupyter server to start. For details, please refer to Installation Guide.
Prerequisites#
Install requirements#
%pip install -q "ultralytics==8.2.24" "matplotlib>=3.4" "onnx<1.16.2" tqdm --extra-index-url https://download.pytorch.org/whl/cpu
%pip install -q "openvino>=2024.4.0"
%pip install -q "nncf>=2.9.0"
%pip install -q "gradio>=4.13"
Note: you may need to restart the kernel to use updated packages.
Note: you may need to restart the kernel to use updated packages.
Note: you may need to restart the kernel to use updated packages.
Note: you may need to restart the kernel to use updated packages.
Imports#
import ipywidgets as widgets
from pathlib import Path
import openvino as ov
import torch
from PIL import Image
from ultralytics import FastSAM
# Fetch skip_kernel_extension module
import requests
r = requests.get(
url="https://raw.githubusercontent.com/openvinotoolkit/openvino_notebooks/latest/utils/skip_kernel_extension.py",
)
open("skip_kernel_extension.py", "w").write(r.text)
# Fetch `notebook_utils` module
import requests
r = requests.get(
url="https://raw.githubusercontent.com/openvinotoolkit/openvino_notebooks/latest/utils/notebook_utils.py",
)
open("notebook_utils.py", "w").write(r.text)
from notebook_utils import download_file, device_widget
%load_ext skip_kernel_extension
FastSAM in Ultralytics#
To work with Fast Segment Anything
Model by
CASIA-IVA-Lab, we will use the Ultralytics
package. Ultralytics package exposes
the FastSAM class, simplifying the model instantiation and weights
loading. The code below demonstrates how to initialize a FastSAM
model and generate a segmentation map.
model_name = "FastSAM-x"
model = FastSAM(model_name)
# Run inference on an image
image_uri = "https://storage.openvinotoolkit.org/repositories/openvino_notebooks/data/data/image/coco_bike.jpg"
image_uri = download_file(image_uri)
results = model(image_uri, device="cpu", retina_masks=True, imgsz=1024, conf=0.6, iou=0.9)
Downloading ultralytics/assets to 'FastSAM-x.pt'...
100%|██████████| 138M/138M [00:03<00:00, 46.3MB/s] /opt/home/k8sworker/ci-ai/cibuilds/jobs/ov-notebook/jobs/OVNotebookOps/builds/835/archive/.workspace/scm/ov-notebook/.venv/lib/python3.8/site-packages/ultralytics/nn/tasks.py:732: FutureWarning: You are using torch.load with weights_only=False (the current default value), which uses the default pickle module implicitly. It is possible to construct malicious pickle data which will execute arbitrary code during unpickling (See pytorch/pytorch for more details). In a future release, the default value for weights_only will be flipped to True. This limits the functions that could be executed during unpickling. Arbitrary objects will no longer be allowed to be loaded via this mode unless they are explicitly allowlisted by the user via torch.serialization.add_safe_globals. We recommend you start setting weights_only=True for any use case where you don't have full control of the loaded file. Please open an issue on GitHub for any issues related to this experimental feature. ckpt = torch.load(file, map_location="cpu")
coco_bike.jpg: 0%| | 0.00/182k [00:00<?, ?B/s]
image 1/1 /opt/home/k8sworker/ci-ai/cibuilds/jobs/ov-notebook/jobs/OVNotebookOps/builds/835/archive/.workspace/scm/ov-notebook/notebooks/fast-segment-anything/coco_bike.jpg: 768x1024 37 objects, 638.3ms
Speed: 3.4ms preprocess, 638.3ms inference, 500.4ms postprocess per image at shape (1, 3, 768, 1024)
The model returns segmentation maps for all the objects on the image. Observe the results below.
Image.fromarray(results[0].plot()[..., ::-1])

Convert the model to OpenVINO Intermediate representation (IR) format#
The Ultralytics Model export API enables conversion of PyTorch models to
OpenVINO IR format. Under the hood it utilizes the
openvino.convert_model method to acquire OpenVINO IR versions of the
models. The method requires a model object and example input for model
tracing. The FastSAM model itself is based on YOLOv8 model.
# instance segmentation model
ov_model_path = Path(f"{model_name}_openvino_model/{model_name}.xml")
if not ov_model_path.exists():
ov_model = model.export(format="openvino", dynamic=False, half=False)
Ultralytics YOLOv8.2.24 🚀 Python-3.8.10 torch-2.4.1+cpu CPU (Intel Core(TM) i9-10920X 3.50GHz)
PyTorch: starting from 'FastSAM-x.pt' with input shape (1, 3, 1024, 1024) BCHW and output shape(s) ((1, 37, 21504), (1, 32, 256, 256)) (138.3 MB)
OpenVINO: starting export with openvino 2024.4.0-16579-c3152d32c9c-releases/2024/4...
OpenVINO: export success ✅ 6.2s, saved as 'FastSAM-x_openvino_model/' (276.1 MB)
Export complete (9.2s)
Results saved to /opt/home/k8sworker/ci-ai/cibuilds/jobs/ov-notebook/jobs/OVNotebookOps/builds/835/archive/.workspace/scm/ov-notebook/notebooks/fast-segment-anything
Predict: yolo predict task=segment model=FastSAM-x_openvino_model imgsz=1024
Validate: yolo val task=segment model=FastSAM-x_openvino_model imgsz=1024 data=ultralytics/datasets/sa.yaml
Visualize: https://netron.app
Embedding the converted models into the original pipeline#
OpenVINO™ Runtime Python API is used to compile the model in OpenVINO IR
format. The
Core
class provides access to the OpenVINO Runtime API. The core object,
which is an instance of the Core class represents the API and it is
used to compile the model.
core = ov.Core()
Select inference device#
Select device that will be used to do models inference using OpenVINO from the dropdown list:
device = device_widget()
device
Dropdown(description='Device:', index=1, options=('CPU', 'AUTO'), value='AUTO')
Adapt OpenVINO models to the original pipeline#
Here we create wrapper classes for the OpenVINO model that we want to
embed in the original inference pipeline. Here are some of the things to
consider when adapting an OV model: - Make sure that parameters passed
by the original pipeline are forwarded to the compiled OV model
properly; sometimes the OV model uses only a portion of the input
arguments and some are ignored, sometimes you need to convert the
argument to another data type or unwrap some data structures such as
tuples or dictionaries. - Guarantee that the wrapper class returns
results to the pipeline in an expected format. In the example below you
can see how we pack OV model outputs into a tuple of torch tensors.
- Pay attention to the model method used in the original pipeline for
calling the model - it may be not the forward method! In this
example, the model is a part of a predictor object and called as and
object, so we need to redefine the magic __call__ method.
class OVWrapper:
def __init__(self, ov_model, device="CPU", stride=32, ov_config=None) -> None:
ov_config = ov_config or {}
self.model = core.compile_model(ov_model, device, ov_config)
self.stride = stride
self.pt = False
self.fp16 = False
self.names = {0: "object"}
def __call__(self, im, **_):
result = self.model(im)
return torch.from_numpy(result[0]), torch.from_numpy(result[1])
Now we initialize the wrapper objects and load them to the FastSAM pipeline.
ov_config = {}
if "GPU" in device.value or ("AUTO" in device.value and "GPU" in core.available_devices):
ov_config = {"GPU_DISABLE_WINOGRAD_CONVOLUTION": "YES"}
wrapped_model = OVWrapper(
ov_model_path,
device=device.value,
stride=model.predictor.model.stride,
ov_config=ov_config,
)
model.predictor.model = wrapped_model
ov_results = model(image_uri, device=device.value, retina_masks=True, imgsz=1024, conf=0.6, iou=0.9)
image 1/1 /opt/home/k8sworker/ci-ai/cibuilds/jobs/ov-notebook/jobs/OVNotebookOps/builds/835/archive/.workspace/scm/ov-notebook/notebooks/fast-segment-anything/coco_bike.jpg: 1024x1024 42 objects, 498.5ms
Speed: 6.1ms preprocess, 498.5ms inference, 31.6ms postprocess per image at shape (1, 3, 1024, 1024)
One can observe the converted model outputs in the next cell, they is the same as of the original model.
Image.fromarray(ov_results[0].plot()[..., ::-1])

Optimize the model using NNCF Post-training Quantization API#
NNCF provides a suite of advanced algorithms for Neural Networks inference optimization in OpenVINO with minimal accuracy drop. We will use 8-bit quantization in post-training mode (without the fine-tuning pipeline) to optimize FastSAM.
The optimization process contains the following steps:
Create a Dataset for quantization.
Run
nncf.quantizeto obtain a quantized model.Save the INT8 model using
openvino.save_model()function.
do_quantize = widgets.Checkbox(
value=True,
description="Quantization",
disabled=False,
)
do_quantize
Checkbox(value=True, description='Quantization')
The nncf.quantize function provides an interface for model
quantization. It requires an instance of the OpenVINO Model and
quantization dataset. Optionally, some additional parameters for the
configuration quantization process (number of samples for quantization,
preset, ignored scope, etc.) can be provided. YOLOv8 model backing
FastSAM contains non-ReLU activation functions, which require asymmetric
quantization of activations. To achieve a better result, we will use a
mixed quantization preset. It provides symmetric quantization of
weights and asymmetric quantization of activations. For more accurate
results, we should keep the operation in the postprocessing subgraph in
floating point precision, using the ignored_scope parameter.
The quantization algorithm is based on The YOLOv8 quantization example in the NNCF repo, refer there for more details. Moreover, you can check out other quantization tutorials in the OV notebooks repo.
Note: Model post-training quantization is time-consuming process. Be patient, it can take several minutes depending on your hardware.
%%skip not $do_quantize.value
import pickle
from contextlib import contextmanager
from zipfile import ZipFile
import cv2
from tqdm.autonotebook import tqdm
import nncf
COLLECT_CALIBRATION_DATA = False
calibration_data = []
@contextmanager
def calibration_data_collection():
global COLLECT_CALIBRATION_DATA
try:
COLLECT_CALIBRATION_DATA = True
yield
finally:
COLLECT_CALIBRATION_DATA = False
class NNCFWrapper:
def __init__(self, ov_model, stride=32) -> None:
self.model = core.read_model(ov_model)
self.compiled_model = core.compile_model(self.model, device_name="CPU")
self.stride = stride
self.pt = False
self.fp16 = False
self.names = {0: "object"}
def __call__(self, im, **_):
if COLLECT_CALIBRATION_DATA:
calibration_data.append(im)
result = self.compiled_model(im)
return torch.from_numpy(result[0]), torch.from_numpy(result[1])
# Fetch data from the web and descibe a dataloader
DATA_URL = "https://ultralytics.com/assets/coco128.zip"
OUT_DIR = Path('.')
download_file(DATA_URL, directory=OUT_DIR, show_progress=True)
if not (OUT_DIR / "coco128/images/train2017").exists():
with ZipFile('coco128.zip', "r") as zip_ref:
zip_ref.extractall(OUT_DIR)
class COCOLoader(torch.utils.data.Dataset):
def __init__(self, images_path):
self.images = list(Path(images_path).iterdir())
def __getitem__(self, index):
if isinstance(index, slice):
return [self.read_image(image_path) for image_path in self.images[index]]
return self.read_image(self.images[index])
def read_image(self, image_path):
image = cv2.imread(str(image_path))
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
return image
def __len__(self):
return len(self.images)
def collect_calibration_data_for_decoder(model, calibration_dataset_size: int,
calibration_cache_path: Path):
global calibration_data
if not calibration_cache_path.exists():
coco_dataset = COCOLoader(OUT_DIR / 'coco128/images/train2017')
with calibration_data_collection():
for image in tqdm(coco_dataset[:calibration_dataset_size], desc="Collecting calibration data"):
model(image, retina_masks=True, imgsz=1024, conf=0.6, iou=0.9, verbose=False)
calibration_cache_path.parent.mkdir(parents=True, exist_ok=True)
with open(calibration_cache_path, "wb") as f:
pickle.dump(calibration_data, f)
else:
with open(calibration_cache_path, "rb") as f:
calibration_data = pickle.load(f)
return calibration_data
def quantize(model, save_model_path: Path, calibration_cache_path: Path,
calibration_dataset_size: int, preset: nncf.QuantizationPreset):
calibration_data = collect_calibration_data_for_decoder(
model, calibration_dataset_size, calibration_cache_path)
quantized_ov_decoder = nncf.quantize(
model.predictor.model.model,
calibration_dataset=nncf.Dataset(calibration_data),
preset=preset,
subset_size=len(calibration_data),
fast_bias_correction=True,
ignored_scope=nncf.IgnoredScope(
types=["Multiply", "Subtract", "Sigmoid"], # ignore operations
names=[
"__module.model.22.dfl.conv/aten::_convolution/Convolution", # in the post-processing subgraph
"__module.model.22/aten::add/Add",
"__module.model.22/aten::add/Add_1"
],
)
)
ov.save_model(quantized_ov_decoder, save_model_path)
wrapped_model = NNCFWrapper(ov_model_path, stride=model.predictor.model.stride)
model.predictor.model = wrapped_model
calibration_dataset_size = 128
quantized_model_path = Path(f"{model_name}_quantized") / "FastSAM-x.xml"
calibration_cache_path = Path(f"calibration_data/coco{calibration_dataset_size}.pkl")
if not quantized_model_path.exists():
quantize(model, quantized_model_path, calibration_cache_path,
calibration_dataset_size=calibration_dataset_size,
preset=nncf.QuantizationPreset.MIXED)
<string>:7: TqdmExperimentalWarning: Using tqdm.autonotebook.tqdm in notebook mode. Use tqdm.tqdm instead to force console mode (e.g. in jupyter console)
INFO:nncf:NNCF initialized successfully. Supported frameworks detected: torch, tensorflow, onnx, openvino
coco128.zip: 0%| | 0.00/6.66M [00:00<?, ?B/s]
Collecting calibration data: 0%| | 0/128 [00:00<?, ?it/s]
INFO:nncf:3 ignored nodes were found by names in the NNCFGraph
INFO:nncf:8 ignored nodes were found by types in the NNCFGraph
INFO:nncf:Not adding activation input quantizer for operation: 268 __module.model.22/aten::sigmoid/Sigmoid
INFO:nncf:Not adding activation input quantizer for operation: 309 __module.model.22.dfl.conv/aten::_convolution/Convolution
INFO:nncf:Not adding activation input quantizer for operation: 346 __module.model.22/aten::sub/Subtract
INFO:nncf:Not adding activation input quantizer for operation: 347 __module.model.22/aten::add/Add
INFO:nncf:Not adding activation input quantizer for operation: 359 __module.model.22/aten::add/Add_1
371 __module.model.22/aten::div/Divide
INFO:nncf:Not adding activation input quantizer for operation: 360 __module.model.22/aten::sub/Subtract_1
INFO:nncf:Not adding activation input quantizer for operation: 382 __module.model.22/aten::mul/Multiply
Output()
Output()
Compare the performance of the Original and Quantized Models#
Finally, we iterate both the OV model and the quantized model over the calibration dataset to measure the performance.
%%skip not $do_quantize.value
import datetime
coco_dataset = COCOLoader(OUT_DIR / 'coco128/images/train2017')
calibration_dataset_size = 128
wrapped_model = OVWrapper(ov_model_path, device=device.value, stride=model.predictor.model.stride)
model.predictor.model = wrapped_model
start_time = datetime.datetime.now()
for image in tqdm(coco_dataset, desc="Measuring inference time"):
model(image, retina_masks=True, imgsz=1024, conf=0.6, iou=0.9, verbose=False)
duration_base = (datetime.datetime.now() - start_time).seconds
print("Segmented in", duration_base, "seconds.")
print("Resulting in", round(calibration_dataset_size / duration_base, 2), "fps")
Measuring inference time: 0%| | 0/128 [00:00<?, ?it/s]
Segmented in 68 seconds.
Resulting in 1.88 fps
%%skip not $do_quantize.value
quantized_wrapped_model = OVWrapper(quantized_model_path, device=device.value, stride=model.predictor.model.stride)
model.predictor.model = quantized_wrapped_model
start_time = datetime.datetime.now()
for image in tqdm(coco_dataset, desc="Measuring inference time"):
model(image, retina_masks=True, imgsz=1024, conf=0.6, iou=0.9, verbose=False)
duration_quantized = (datetime.datetime.now() - start_time).seconds
print("Segmented in", duration_quantized, "seconds")
print("Resulting in", round(calibration_dataset_size / duration_quantized, 2), "fps")
print("That is", round(duration_base / duration_quantized, 2), "times faster!")
Measuring inference time: 0%| | 0/128 [00:00<?, ?it/s]
Segmented in 21 seconds
Resulting in 6.1 fps
That is 3.24 times faster!
Try out the converted pipeline#
The demo app below is created using Gradio package.
The app allows you to alter the model output interactively. Using the Pixel selector type switch you can place foreground/background points or bounding boxes on input image.
import cv2
import numpy as np
import matplotlib.pyplot as plt
def fast_process(
annotations,
image,
scale,
better_quality=False,
mask_random_color=True,
bbox=None,
use_retina=True,
with_contours=True,
):
original_h = image.height
original_w = image.width
if better_quality:
for i, mask in enumerate(annotations):
mask = cv2.morphologyEx(mask.astype(np.uint8), cv2.MORPH_CLOSE, np.ones((3, 3), np.uint8))
annotations[i] = cv2.morphologyEx(mask.astype(np.uint8), cv2.MORPH_OPEN, np.ones((8, 8), np.uint8))
inner_mask = fast_show_mask(
annotations,
plt.gca(),
random_color=mask_random_color,
bbox=bbox,
retinamask=use_retina,
target_height=original_h,
target_width=original_w,
)
if with_contours:
contour_all = []
temp = np.zeros((original_h, original_w, 1))
for i, mask in enumerate(annotations):
annotation = mask.astype(np.uint8)
if not use_retina:
annotation = cv2.resize(
annotation,
(original_w, original_h),
interpolation=cv2.INTER_NEAREST,
)
contours, _ = cv2.findContours(annotation, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
for contour in contours:
contour_all.append(contour)
cv2.drawContours(temp, contour_all, -1, (255, 255, 255), 2 // scale)
color = np.array([0 / 255, 0 / 255, 255 / 255, 0.9])
contour_mask = temp / 255 * color.reshape(1, 1, -1)
image = image.convert("RGBA")
overlay_inner = Image.fromarray((inner_mask * 255).astype(np.uint8), "RGBA")
image.paste(overlay_inner, (0, 0), overlay_inner)
if with_contours:
overlay_contour = Image.fromarray((contour_mask * 255).astype(np.uint8), "RGBA")
image.paste(overlay_contour, (0, 0), overlay_contour)
return image
# CPU post process
def fast_show_mask(
annotation,
ax,
random_color=False,
bbox=None,
retinamask=True,
target_height=960,
target_width=960,
):
mask_sum = annotation.shape[0]
height = annotation.shape[1]
weight = annotation.shape[2]
#
areas = np.sum(annotation, axis=(1, 2))
sorted_indices = np.argsort(areas)[::1]
annotation = annotation[sorted_indices]
index = (annotation != 0).argmax(axis=0)
if random_color:
color = np.random.random((mask_sum, 1, 1, 3))
else:
color = np.ones((mask_sum, 1, 1, 3)) * np.array([30 / 255, 144 / 255, 255 / 255])
transparency = np.ones((mask_sum, 1, 1, 1)) * 0.6
visual = np.concatenate([color, transparency], axis=-1)
mask_image = np.expand_dims(annotation, -1) * visual
mask = np.zeros((height, weight, 4))
h_indices, w_indices = np.meshgrid(np.arange(height), np.arange(weight), indexing="ij")
indices = (index[h_indices, w_indices], h_indices, w_indices, slice(None))
mask[h_indices, w_indices, :] = mask_image[indices]
if bbox is not None:
x1, y1, x2, y2 = bbox
ax.add_patch(plt.Rectangle((x1, y1), x2 - x1, y2 - y1, fill=False, edgecolor="b", linewidth=1))
if not retinamask:
mask = cv2.resize(mask, (target_width, target_height), interpolation=cv2.INTER_NEAREST)
return mask
This is the main callback function that is called to segment an image based on user input.
object_points = []
background_points = []
bbox_points = []
def segment(
image,
model_type,
input_size=1024,
iou_threshold=0.75,
conf_threshold=0.4,
better_quality=True,
with_contours=True,
use_retina=True,
mask_random_color=True,
):
if do_quantize.value and model_type == "Quantized model":
model.predictor.model = quantized_wrapped_model
else:
model.predictor.model = wrapped_model
input_size = int(input_size)
w, h = image.size
scale = input_size / max(w, h)
new_w = int(w * scale)
new_h = int(h * scale)
image = image.resize((new_w, new_h))
results = model(
image,
retina_masks=use_retina,
iou=iou_threshold,
conf=conf_threshold,
imgsz=input_size,
)
masks = results[0].masks.data
# Calculate annotations
if not (object_points or bbox_points):
annotations = masks.cpu().numpy()
else:
annotations = []
if object_points:
all_points = object_points + background_points
labels = [1] * len(object_points) + [0] * len(background_points)
scaled_points = [[int(x * scale) for x in point] for point in all_points]
h, w = masks[0].shape[:2]
assert max(h, w) == input_size
onemask = np.zeros((h, w))
for mask in sorted(masks, key=lambda x: x.sum(), reverse=True):
mask_np = (mask == 1.0).cpu().numpy()
for point, label in zip(scaled_points, labels):
if mask_np[point[1], point[0]] == 1 and label == 1:
onemask[mask_np] = 1
if mask_np[point[1], point[0]] == 1 and label == 0:
onemask[mask_np] = 0
annotations.append(onemask >= 1)
if len(bbox_points) >= 2:
scaled_bbox_points = []
for i, point in enumerate(bbox_points):
x, y = int(point[0] * scale), int(point[1] * scale)
x = max(min(x, new_w), 0)
y = max(min(y, new_h), 0)
scaled_bbox_points.append((x, y))
for i in range(0, len(scaled_bbox_points) - 1, 2):
x0, y0, x1, y1 = *scaled_bbox_points[i], *scaled_bbox_points[i + 1]
intersection_area = torch.sum(masks[:, y0:y1, x0:x1], dim=(1, 2))
masks_area = torch.sum(masks, dim=(1, 2))
bbox_area = (y1 - y0) * (x1 - x0)
union = bbox_area + masks_area - intersection_area
iou = intersection_area / union
max_iou_index = torch.argmax(iou)
annotations.append(masks[max_iou_index].cpu().numpy())
return fast_process(
annotations=np.array(annotations),
image=image,
scale=(1024 // input_size),
better_quality=better_quality,
mask_random_color=mask_random_color,
bbox=None,
use_retina=use_retina,
with_contours=with_contours,
)
if not Path("gradio_helper.py").exists():
r = requests.get(url="https://raw.githubusercontent.com/openvinotoolkit/openvino_notebooks/latest/notebooks/fast-segment-anything/gradio_helper.py")
open("gradio_helper.py", "w").write(r.text)
from gradio_helper import make_demo
demo = make_demo(fn=segment, quantized=do_quantize.value)
try:
demo.queue().launch(debug=False)
except Exception:
demo.queue().launch(share=True, debug=False)
# If you are launching remotely, specify server_name and server_port
# EXAMPLE: `demo.launch(server_name="your server name", server_port="server port in int")`
# To learn more please refer to the Gradio docs: https://gradio.app/docs/
Running on local URL: http://127.0.0.1:7860 To create a public link, set share=True in launch().