Quantize a Segmentation Model and Show Live Inference¶
This Jupyter notebook can be launched after a local installation only.
Kidney Segmentation with PyTorch Lightning and OpenVINO™ - Part 3¶
This tutorial is a part of a series on how to train, optimize, quantize and show live inference on a medical segmentation model. The goal is to accelerate inference on a kidney segmentation model. The UNet model is trained from scratch; the data is from Kits19.
This third tutorial in the series shows how to:
Convert an Original model to OpenVINO IR with model conversion API
Quantize a PyTorch model with NNCF
Evaluate the F1 score metric of the original model and the quantized model
Benchmark performance of the FP32 model and the INT8 quantized model
Show live inference with OpenVINO’s async API
All notebooks in this series:
Train a 2D-UNet Medical Imaging Model with PyTorch Lightning
Convert and Quantize a Segmentation Model and Show Live Inference (this notebook)
Instructions¶
This notebook needs a trained UNet model. We provide a pre-trained model, trained for 20 epochs with the full Kits-19 frames dataset, which has an F1 score on the validation set of 0.9. The training code is available in this notebook.
NNCF for PyTorch models requires a C++ compiler. On Windows, install
Microsoft Visual Studio
2019.
During installation, choose Desktop development with C++ in the
Workloads tab. On macOS, run xcode-select –install from a Terminal.
On Linux, install gcc.
Running this notebook with the full dataset will take a long time. For demonstration purposes, this tutorial will download one converted CT scan and use that scan for quantization and inference. For production purposes, use a representative dataset for quantizing the model.
Table of contents:¶
%pip install -q "openvino>=2023.1.0" "monai>=0.9.1,<1.0.0" "torchmetrics>=0.11.0" "nncf>=2.6.0" --extra-index-url https://download.pytorch.org/whl/cpu
Note: you may need to restart the kernel to use updated packages.
Imports¶
# On Windows, try to find the directory that contains x64 cl.exe and add it to the PATH to enable PyTorch
# to find the required C++ tools. This code assumes that Visual Studio is installed in the default
# directory. If you have a different C++ compiler, please add the correct path to os.environ["PATH"]
# directly. Note that the C++ Redistributable is not enough to run this notebook.
# Adding the path to os.environ["LIB"] is not always required - it depends on the system's configuration
import sys
if sys.platform == "win32":
import distutils.command.build_ext
import os
from pathlib import Path
if sys.getwindowsversion().build >= 20000: # Windows 11
search_path = "**/Hostx64/x64/cl.exe"
else:
search_path = "**/Hostx86/x64/cl.exe"
VS_INSTALL_DIR_2019 = r"C:/Program Files (x86)/Microsoft Visual Studio"
VS_INSTALL_DIR_2022 = r"C:/Program Files/Microsoft Visual Studio"
cl_paths_2019 = sorted(list(Path(VS_INSTALL_DIR_2019).glob(search_path)))
cl_paths_2022 = sorted(list(Path(VS_INSTALL_DIR_2022).glob(search_path)))
cl_paths = cl_paths_2019 + cl_paths_2022
if len(cl_paths) == 0:
raise ValueError(
"Cannot find Visual Studio. This notebook requires an x64 C++ compiler. If you installed "
"a C++ compiler, please add the directory that contains cl.exe to `os.environ['PATH']`."
)
else:
# If multiple versions of MSVC are installed, get the most recent version
cl_path = cl_paths[-1]
vs_dir = str(cl_path.parent)
os.environ["PATH"] += f"{os.pathsep}{vs_dir}"
# Code for finding the library dirs from
# https://stackoverflow.com/questions/47423246/get-pythons-lib-path
d = distutils.core.Distribution()
b = distutils.command.build_ext.build_ext(d)
b.finalize_options()
os.environ["LIB"] = os.pathsep.join(b.library_dirs)
print(f"Added {vs_dir} to PATH")
import logging
import os
import random
import sys
import time
import warnings
import zipfile
from pathlib import Path
from typing import Union
warnings.filterwarnings("ignore", category=UserWarning)
import cv2
import matplotlib.pyplot as plt
import monai
import numpy as np
import torch
import nncf
import openvino as ov
from monai.transforms import LoadImage
from nncf.common.logging.logger import set_log_level
from torchmetrics import F1Score as F1
set_log_level(logging.ERROR) # Disables all NNCF info and warning messages
from custom_segmentation import SegmentationModel
from async_pipeline import show_live_inference
sys.path.append("../utils")
from notebook_utils import download_file
2024-02-09 22:51:11.599112: I tensorflow/core/util/port.cc:110] oneDNN custom operations are on. You may see slightly different numerical results due to floating-point round-off errors from different computation orders. To turn them off, set the environment variable TF_ENABLE_ONEDNN_OPTS=0. 2024-02-09 22:51:11.634091: I tensorflow/core/platform/cpu_feature_guard.cc:182] This TensorFlow binary is optimized to use available CPU instructions in performance-critical operations. To enable the following instructions: AVX2 AVX512F AVX512_VNNI FMA, in other operations, rebuild TensorFlow with the appropriate compiler flags.
2024-02-09 22:51:12.223414: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Could not find TensorRT
INFO:nncf:NNCF initialized successfully. Supported frameworks detected: torch, tensorflow, onnx, openvino
Settings¶
By default, this notebook will download one CT scan from the KITS19
dataset that will be used for quantization. To use the full dataset, set
BASEDIR to the path of the dataset, as prepared according to the
Data Preparation notebook.
BASEDIR = Path("kits19_frames_1")
# Uncomment the line below to use the full dataset, as prepared in the data preparation notebook
# BASEDIR = Path("~/kits19/kits19_frames").expanduser()
MODEL_DIR = Path("model")
MODEL_DIR.mkdir(exist_ok=True)
Load PyTorch Model¶
Download the pre-trained model weights, load the PyTorch model and the
state_dict that was saved after training. The model used in this
notebook is a
BasicUNet
model from MONAI. We provide a pre-trained
checkpoint. To see how this model performs, check out the training
notebook.
state_dict_url = "https://storage.openvinotoolkit.org/repositories/openvino_notebooks/models/kidney-segmentation-kits19/unet_kits19_state_dict.pth"
state_dict_file = download_file(state_dict_url, directory="pretrained_model")
state_dict = torch.load(state_dict_file, map_location=torch.device("cpu"))
new_state_dict = {}
for k, v in state_dict.items():
new_key = k.replace("_model.", "")
new_state_dict[new_key] = v
new_state_dict.pop("loss_function.pos_weight")
model = monai.networks.nets.BasicUNet(spatial_dims=2, in_channels=1, out_channels=1).eval()
model.load_state_dict(new_state_dict)
pretrained_model/unet_kits19_state_dict.pth: 0%| | 0.00/7.58M [00:00<?, ?B/s]
BasicUNet features: (32, 32, 64, 128, 256, 32).
<All keys matched successfully>
Download CT-scan Data¶
# The CT scan case number. For example: 2 for data from the case_00002 directory
# Currently only 117 is supported
CASE = 117
if not (BASEDIR / f"case_{CASE:05d}").exists():
BASEDIR.mkdir(exist_ok=True)
filename = download_file(
f"https://storage.openvinotoolkit.org/data/test_data/openvino_notebooks/kits19/case_{CASE:05d}.zip"
)
with zipfile.ZipFile(filename, "r") as zip_ref:
zip_ref.extractall(path=BASEDIR)
os.remove(filename) # remove zipfile
print(f"Downloaded and extracted data for case_{CASE:05d}")
else:
print(f"Data for case_{CASE:05d} exists")
Data for case_00117 exists
Configuration¶
The KitsDataset class in the next cell expects images and masks in
the ``basedir`` directory, in a folder per patient. It is a simplified
version of the Dataset class in the training
notebook.
Images are loaded with MONAI’s
LoadImage,
to align with the image loading method in the training notebook. This
method rotates and flips the images. We define a rotate_and_flip
method to display the images in the expected orientation:
def rotate_and_flip(image):
"""Rotate `image` by 90 degrees and flip horizontally"""
return cv2.flip(cv2.rotate(image, rotateCode=cv2.ROTATE_90_CLOCKWISE), flipCode=1)
class KitsDataset:
def __init__(self, basedir: str):
"""
Dataset class for prepared Kits19 data, for binary segmentation (background/kidney)
Source data should exist in basedir, in subdirectories case_00000 until case_00210,
with each subdirectory containing directories imaging_frames, with jpg images, and
segmentation_frames with segmentation masks as png files.
See https://github.com/openvinotoolkit/openvino_notebooks/blob/main/notebooks/110-ct-segmentation-quantize/data-preparation-ct-scan.ipynb
:param basedir: Directory that contains the prepared CT scans
"""
masks = sorted(BASEDIR.glob("case_*/segmentation_frames/*png"))
self.basedir = basedir
self.dataset = masks
print(
f"Created dataset with {len(self.dataset)} items. "
f"Base directory for data: {basedir}"
)
def __getitem__(self, index):
"""
Get an item from the dataset at the specified index.
:return: (image, segmentation_mask)
"""
mask_path = self.dataset[index]
image_path = str(mask_path.with_suffix(".jpg")).replace(
"segmentation_frames", "imaging_frames"
)
# Load images with MONAI's LoadImage to match data loading in training notebook
mask = LoadImage(image_only=True, dtype=np.uint8)(str(mask_path)).numpy()
img = LoadImage(image_only=True, dtype=np.float32)(str(image_path)).numpy()
if img.shape[:2] != (512, 512):
img = cv2.resize(img.astype(np.uint8), (512, 512)).astype(np.float32)
mask = cv2.resize(mask, (512, 512))
input_image = np.expand_dims(img, axis=0)
return input_image, mask
def __len__(self):
return len(self.dataset)
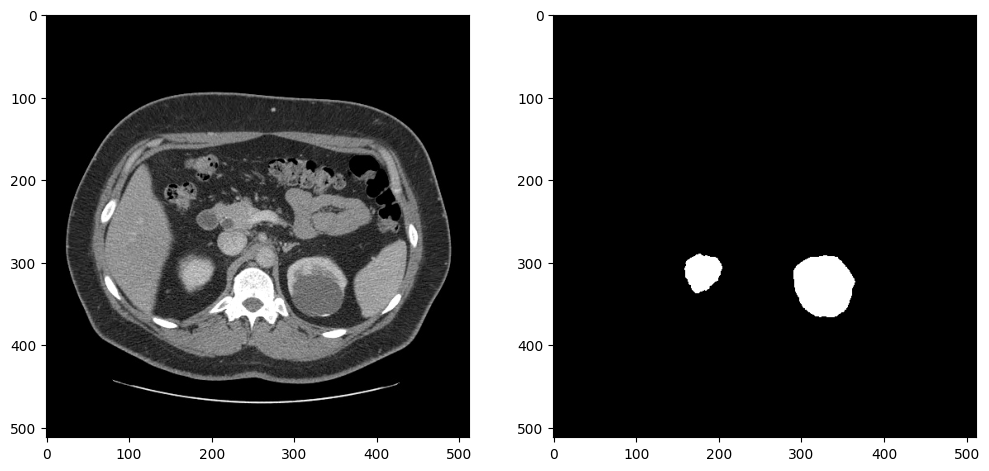
To test whether the data loader returns the expected output, we show an image and a mask. The image and the mask are returned by the dataloader, after resizing and preprocessing. Since this dataset contains a lot of slices without kidneys, we select a slice that contains at least 5000 kidney pixels to verify that the annotations look correct:
dataset = KitsDataset(BASEDIR)
# Find a slice that contains kidney annotations
# item[0] is the annotation: (id, annotation_data)
image_data, mask = next(item for item in dataset if np.count_nonzero(item[1]) > 5000)
# Remove extra image dimension and rotate and flip the image for visualization
image = rotate_and_flip(image_data.squeeze())
# The data loader returns annotations as (index, mask) and mask in shape (H,W)
mask = rotate_and_flip(mask)
fig, ax = plt.subplots(1, 2, figsize=(12, 6))
ax[0].imshow(image, cmap="gray")
ax[1].imshow(mask, cmap="gray");
Created dataset with 69 items. Base directory for data: kits19_frames_1
Define a metric to determine the performance of the model.
For this demo, we use the F1 score, or Dice coefficient, from the TorchMetrics library.
def compute_f1(model: Union[torch.nn.Module, ov.CompiledModel], dataset: KitsDataset):
"""
Compute binary F1 score of `model` on `dataset`
F1 score metric is provided by the torchmetrics library
`model` is expected to be a binary segmentation model, images in the
dataset are expected in (N,C,H,W) format where N==C==1
"""
metric = F1(ignore_index=0, task="binary", average="macro")
with torch.no_grad():
for image, target in dataset:
input_image = torch.as_tensor(image).unsqueeze(0)
if isinstance(model, ov.CompiledModel):
output_layer = model.output(0)
output = model(input_image)[output_layer]
output = torch.from_numpy(output)
else:
output = model(input_image)
label = torch.as_tensor(target.squeeze()).long()
prediction = torch.sigmoid(output.squeeze()).round().long()
metric.update(label.flatten(), prediction.flatten())
return metric.compute()
Quantization¶
Before quantizing the model, we compute the F1 score on the FP32
model, for comparison:
fp32_f1 = compute_f1(model, dataset)
print(f"FP32 F1: {fp32_f1:.3f}")
FP32 F1: 0.999
We convert the PyTorch model to OpenVINO IR and serialize it for
comparing the performance of the FP32 and INT8 model later in
this notebook.
fp32_ir_path = MODEL_DIR / Path('unet_kits19_fp32.xml')
fp32_ir_model = ov.convert_model(model, example_input=torch.ones(1, 1, 512, 512, dtype=torch.float32))
ov.save_model(fp32_ir_model, str(fp32_ir_path))
WARNING:tensorflow:Please fix your imports. Module tensorflow.python.training.tracking.base has been moved to tensorflow.python.trackable.base. The old module will be deleted in version 2.11.
[ WARNING ] Please fix your imports. Module %s has been moved to %s. The old module will be deleted in version %s.
No CUDA runtime is found, using CUDA_HOME='/usr/local/cuda'
/opt/home/k8sworker/ci-ai/cibuilds/ov-notebook/OVNotebookOps-609/.workspace/scm/ov-notebook/.venv/lib/python3.8/site-packages/monai/networks/nets/basic_unet.py:179: TracerWarning: Converting a tensor to a Python boolean might cause the trace to be incorrect. We can't record the data flow of Python values, so this value will be treated as a constant in the future. This means that the trace might not generalize to other inputs!
if x_e.shape[-i - 1] != x_0.shape[-i - 1]:
NNCF provides a suite of advanced algorithms for Neural Networks inference optimization in OpenVINO with minimal accuracy drop.
NOTE: NNCF Post-training Quantization is available in OpenVINO 2023.0 release.
Create a quantized model from the pre-trained FP32 model and the
calibration dataset. The optimization process contains the following
steps:
1. Create a Dataset for quantization.
2. Run `nncf.quantize` for getting an optimized model.
3. Export the quantized model to ONNX and then convert to OpenVINO IR model.
4. Serialize the INT8 model using `ov.save_model` function for benchmarking.
def transform_fn(data_item):
"""
Extract the model's input from the data item.
The data item here is the data item that is returned from the data source per iteration.
This function should be passed when the data item cannot be used as model's input.
"""
images, _ = data_item
return images
data_loader = torch.utils.data.DataLoader(dataset)
calibration_dataset = nncf.Dataset(data_loader, transform_fn)
quantized_model = nncf.quantize(
model,
calibration_dataset,
# Do not quantize LeakyReLU activations to allow the INT8 model to run on Intel GPU
ignored_scope=nncf.IgnoredScope(patterns=[".*LeakyReLU.*"])
)
Output()
Output()
Export the quantized model to ONNX and then convert it to OpenVINO IR model and save it.
dummy_input = torch.randn(1, 1, 512, 512)
int8_onnx_path = MODEL_DIR / "unet_kits19_int8.onnx"
int8_ir_path = Path(int8_onnx_path).with_suffix(".xml")
torch.onnx.export(quantized_model, dummy_input, int8_onnx_path)
int8_ir_model = ov.convert_model(int8_onnx_path)
ov.save_model(int8_ir_model, str(int8_ir_path))
/opt/home/k8sworker/ci-ai/cibuilds/ov-notebook/OVNotebookOps-609/.workspace/scm/ov-notebook/.venv/lib/python3.8/site-packages/nncf/torch/quantization/layers.py:334: TracerWarning: Converting a tensor to a Python number might cause the trace to be incorrect. We can't record the data flow of Python values, so this value will be treated as a constant in the future. This means that the trace might not generalize to other inputs!
return self._level_low.item()
/opt/home/k8sworker/ci-ai/cibuilds/ov-notebook/OVNotebookOps-609/.workspace/scm/ov-notebook/.venv/lib/python3.8/site-packages/nncf/torch/quantization/layers.py:342: TracerWarning: Converting a tensor to a Python number might cause the trace to be incorrect. We can't record the data flow of Python values, so this value will be treated as a constant in the future. This means that the trace might not generalize to other inputs!
return self._level_high.item()
/opt/home/k8sworker/ci-ai/cibuilds/ov-notebook/OVNotebookOps-609/.workspace/scm/ov-notebook/.venv/lib/python3.8/site-packages/monai/networks/nets/basic_unet.py:179: TracerWarning: Converting a tensor to a Python boolean might cause the trace to be incorrect. We can't record the data flow of Python values, so this value will be treated as a constant in the future. This means that the trace might not generalize to other inputs!
if x_e.shape[-i - 1] != x_0.shape[-i - 1]:
This notebook demonstrates post-training quantization with NNCF.
NNCF also supports quantization-aware training, and other algorithms than quantization. See the NNCF documentation in the NNCF repository for more information.
Compare FP32 and INT8 Model¶
fp32_ir_model_size = fp32_ir_path.with_suffix(".bin").stat().st_size / 1024
quantized_model_size = int8_ir_path.with_suffix(".bin").stat().st_size / 1024
print(f"FP32 IR model size: {fp32_ir_model_size:.2f} KB")
print(f"INT8 model size: {quantized_model_size:.2f} KB")
FP32 IR model size: 3864.14 KB
INT8 model size: 1940.41 KB
core = ov.Core()
int8_compiled_model = core.compile_model(int8_ir_model)
int8_f1 = compute_f1(int8_compiled_model, dataset)
print(f"FP32 F1: {fp32_f1:.3f}")
print(f"INT8 F1: {int8_f1:.3f}")
FP32 F1: 0.999
INT8 F1: 0.999
To measure the inference performance of the FP32 and INT8
models, we use Benchmark
Tool
- OpenVINO’s inference performance measurement tool. Benchmark tool is a
command line application, part of OpenVINO development tools, that can
be run in the notebook with ! benchmark_app or
%sx benchmark_app.
NOTE: For the most accurate performance estimation, it is recommended to run
benchmark_appin a terminal/command prompt after closing other applications. Runbenchmark_app -m model.xml -d CPUto benchmark async inference on CPU for one minute. ChangeCPUtoGPUto benchmark on GPU. Runbenchmark_app --helpto see all command line options.
# ! benchmark_app --help
device = "CPU"
# Benchmark FP32 model
! benchmark_app -m $fp32_ir_path -d $device -t 15 -api sync
[Step 1/11] Parsing and validating input arguments
[ INFO ] Parsing input parameters
[Step 2/11] Loading OpenVINO Runtime
[ INFO ] OpenVINO:
[ INFO ] Build ................................. 2023.3.0-13775-ceeafaf64f3-releases/2023/3
[ INFO ]
[ INFO ] Device info:
[ INFO ] CPU [ INFO ] Build ................................. 2023.3.0-13775-ceeafaf64f3-releases/2023/3 [ INFO ] [ INFO ] [Step 3/11] Setting device configuration [ WARNING ] Performance hint was not explicitly specified in command line. Device(CPU) performance hint will be set to PerformanceMode.LATENCY. [Step 4/11] Reading model files [ INFO ] Loading model files [ INFO ] Read model took 26.51 ms [ INFO ] Original model I/O parameters: [ INFO ] Model inputs: [ INFO ] x (node: x) : f32 / [...] / [?,?,?,?] [ INFO ] Model outputs: [ INFO ] *NO_NAME* (node: __module.final_conv/aten::_convolution/Add) : f32 / [...] / [?,1,16..,16..] [Step 5/11] Resizing model to match image sizes and given batch [ INFO ] Model batch size: 1 [Step 6/11] Configuring input of the model [ INFO ] Model inputs: [ INFO ] x (node: x) : f32 / [...] / [?,?,?,?] [ INFO ] Model outputs: [ INFO ] *NO_NAME* (node: __module.final_conv/aten::_convolution/Add) : f32 / [...] / [?,1,16..,16..] [Step 7/11] Loading the model to the device
[ INFO ] Compile model took 87.61 ms
[Step 8/11] Querying optimal runtime parameters
[ INFO ] Model:
[ INFO ] NETWORK_NAME: Model0
[ INFO ] OPTIMAL_NUMBER_OF_INFER_REQUESTS: 1
[ INFO ] NUM_STREAMS: 1
[ INFO ] AFFINITY: Affinity.CORE
[ INFO ] INFERENCE_NUM_THREADS: 12
[ INFO ] PERF_COUNT: NO
[ INFO ] INFERENCE_PRECISION_HINT: <Type: 'float32'>
[ INFO ] PERFORMANCE_HINT: LATENCY
[ INFO ] EXECUTION_MODE_HINT: ExecutionMode.PERFORMANCE
[ INFO ] PERFORMANCE_HINT_NUM_REQUESTS: 0
[ INFO ] ENABLE_CPU_PINNING: True
[ INFO ] SCHEDULING_CORE_TYPE: SchedulingCoreType.ANY_CORE
[ INFO ] ENABLE_HYPER_THREADING: False
[ INFO ] EXECUTION_DEVICES: ['CPU']
[ INFO ] CPU_DENORMALS_OPTIMIZATION: False
[ INFO ] CPU_SPARSE_WEIGHTS_DECOMPRESSION_RATE: 1.0
[Step 9/11] Creating infer requests and preparing input tensors
[ ERROR ] Input x is dynamic. Provide data shapes!
Traceback (most recent call last):
File "/opt/home/k8sworker/ci-ai/cibuilds/ov-notebook/OVNotebookOps-609/.workspace/scm/ov-notebook/.venv/lib/python3.8/site-packages/openvino/tools/benchmark/main.py", line 486, in main
data_queue = get_input_data(paths_to_input, app_inputs_info)
File "/opt/home/k8sworker/ci-ai/cibuilds/ov-notebook/OVNotebookOps-609/.workspace/scm/ov-notebook/.venv/lib/python3.8/site-packages/openvino/tools/benchmark/utils/inputs_filling.py", line 123, in get_input_data
raise Exception(f"Input {info.name} is dynamic. Provide data shapes!")
Exception: Input x is dynamic. Provide data shapes!
# Benchmark INT8 model
! benchmark_app -m $int8_ir_path -d $device -t 15 -api sync
[Step 1/11] Parsing and validating input arguments
[ INFO ] Parsing input parameters
[Step 2/11] Loading OpenVINO Runtime
[ INFO ] OpenVINO:
[ INFO ] Build ................................. 2023.3.0-13775-ceeafaf64f3-releases/2023/3
[ INFO ]
[ INFO ] Device info:
[ INFO ] CPU
[ INFO ] Build ................................. 2023.3.0-13775-ceeafaf64f3-releases/2023/3
[ INFO ]
[ INFO ]
[Step 3/11] Setting device configuration
[ WARNING ] Performance hint was not explicitly specified in command line. Device(CPU) performance hint will be set to PerformanceMode.LATENCY.
[Step 4/11] Reading model files
[ INFO ] Loading model files
[ INFO ] Read model took 13.17 ms
[ INFO ] Original model I/O parameters:
[ INFO ] Model inputs:
[ INFO ] x.1 (node: x.1) : f32 / [...] / [1,1,512,512]
[ INFO ] Model outputs:
[ INFO ] 571 (node: 571) : f32 / [...] / [1,1,512,512]
[Step 5/11] Resizing model to match image sizes and given batch
[ INFO ] Model batch size: 1
[Step 6/11] Configuring input of the model
[ INFO ] Model inputs:
[ INFO ] x.1 (node: x.1) : f32 / [N,C,H,W] / [1,1,512,512]
[ INFO ] Model outputs:
[ INFO ] 571 (node: 571) : f32 / [...] / [1,1,512,512]
[Step 7/11] Loading the model to the device
[ INFO ] Compile model took 190.56 ms
[Step 8/11] Querying optimal runtime parameters
[ INFO ] Model:
[ INFO ] NETWORK_NAME: main_graph
[ INFO ] OPTIMAL_NUMBER_OF_INFER_REQUESTS: 1
[ INFO ] NUM_STREAMS: 1
[ INFO ] AFFINITY: Affinity.CORE
[ INFO ] INFERENCE_NUM_THREADS: 12
[ INFO ] PERF_COUNT: NO
[ INFO ] INFERENCE_PRECISION_HINT: <Type: 'float32'>
[ INFO ] PERFORMANCE_HINT: LATENCY
[ INFO ] EXECUTION_MODE_HINT: ExecutionMode.PERFORMANCE
[ INFO ] PERFORMANCE_HINT_NUM_REQUESTS: 0
[ INFO ] ENABLE_CPU_PINNING: True
[ INFO ] SCHEDULING_CORE_TYPE: SchedulingCoreType.ANY_CORE
[ INFO ] ENABLE_HYPER_THREADING: False
[ INFO ] EXECUTION_DEVICES: ['CPU']
[ INFO ] CPU_DENORMALS_OPTIMIZATION: False
[ INFO ] CPU_SPARSE_WEIGHTS_DECOMPRESSION_RATE: 1.0
[Step 9/11] Creating infer requests and preparing input tensors
[ WARNING ] No input files were given for input 'x.1'!. This input will be filled with random values!
[ INFO ] Fill input 'x.1' with random values
[Step 10/11] Measuring performance (Start inference synchronously, limits: 15000 ms duration)
[ INFO ] Benchmarking in inference only mode (inputs filling are not included in measurement loop).
[ INFO ] First inference took 30.25 ms
[Step 11/11] Dumping statistics report
[ INFO ] Execution Devices:['CPU']
[ INFO ] Count: 973 iterations
[ INFO ] Duration: 15006.05 ms
[ INFO ] Latency:
[ INFO ] Median: 15.16 ms
[ INFO ] Average: 15.22 ms
[ INFO ] Min: 14.87 ms
[ INFO ] Max: 17.88 ms
[ INFO ] Throughput: 64.84 FPS
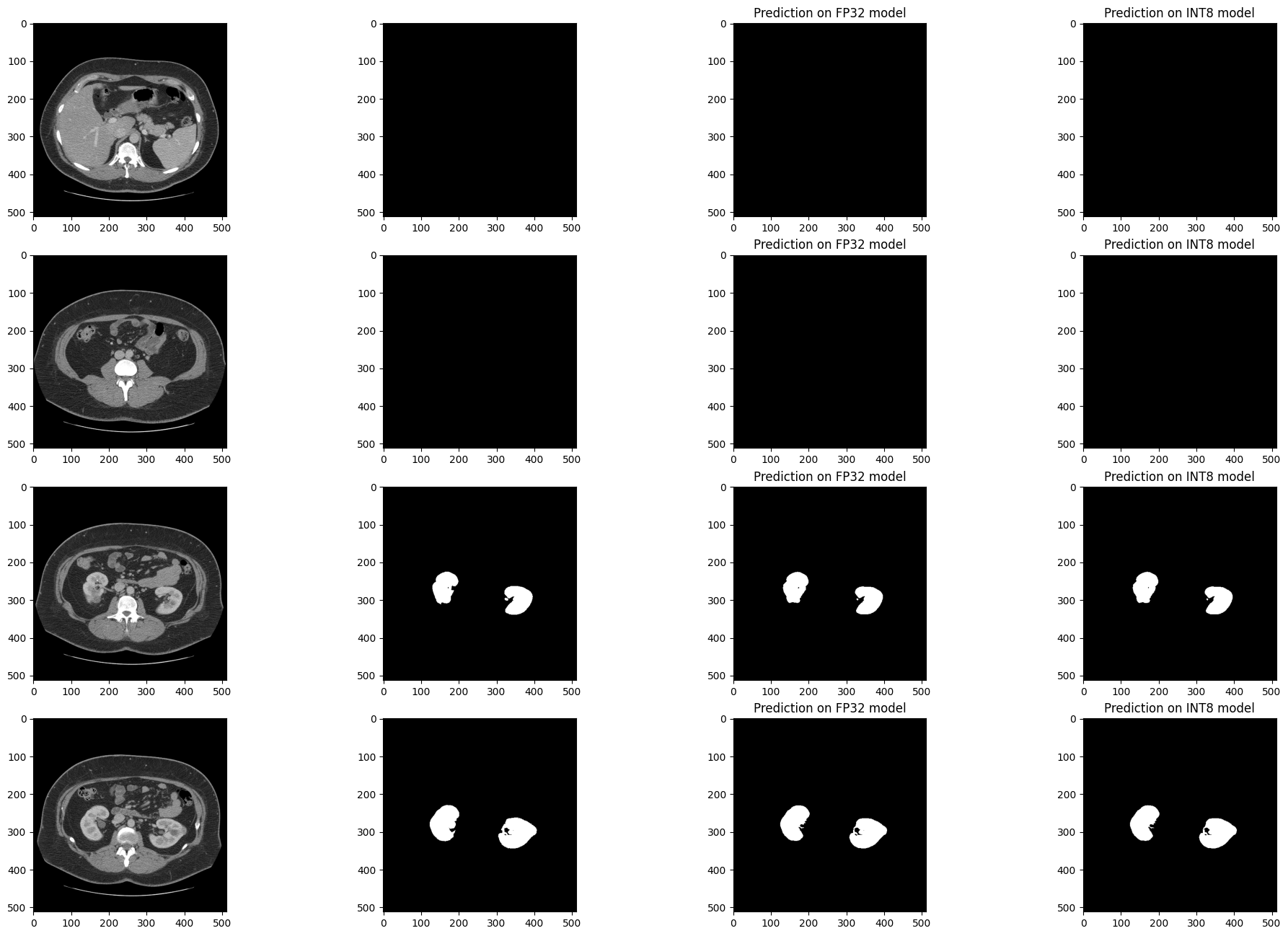
Visualize the results of the model on four slices of the validation set.
Compare the results of the FP32 IR model with the results of the
quantized INT8 model and the reference segmentation annotation.
Medical imaging datasets tend to be very imbalanced: most of the slices in a CT scan do not contain kidney data. The segmentation model should be good at finding kidneys where they exist (in medical terms: have good sensitivity) but also not find spurious kidneys that do not exist (have good specificity). In the next cell, there are four slices: two slices that have no kidney data, and two slices that contain kidney data. For this example, a slice has kidney data if at least 50 pixels in the slices are annotated as kidney.
Run this cell again to show results on a different subset. The random seed is displayed to enable reproducing specific runs of this cell.
NOTE: the images are shown after optional augmenting and resizing. In the Kits19 dataset all but one of the cases has the
(512, 512)input shape.
# The sigmoid function is used to transform the result of the network
# to binary segmentation masks
def sigmoid(x):
return np.exp(-np.logaddexp(0, -x))
num_images = 4
colormap = "gray"
# Load FP32 and INT8 models
core = ov.Core()
fp_model = core.read_model(fp32_ir_path)
int8_model = core.read_model(int8_ir_path)
compiled_model_fp = core.compile_model(fp_model, device_name="CPU")
compiled_model_int8 = core.compile_model(int8_model, device_name="CPU")
output_layer_fp = compiled_model_fp.output(0)
output_layer_int8 = compiled_model_int8.output(0)
# Create subset of dataset
background_slices = (item for item in dataset if np.count_nonzero(item[1]) == 0)
kidney_slices = (item for item in dataset if np.count_nonzero(item[1]) > 50)
data_subset = random.sample(list(background_slices), 2) + random.sample(list(kidney_slices), 2)
# Set seed to current time. To reproduce specific results, copy the printed seed
# and manually set `seed` to that value.
seed = int(time.time())
random.seed(seed)
print(f"Visualizing results with seed {seed}")
fig, ax = plt.subplots(nrows=num_images, ncols=4, figsize=(24, num_images * 4))
for i, (image, mask) in enumerate(data_subset):
display_image = rotate_and_flip(image.squeeze())
target_mask = rotate_and_flip(mask).astype(np.uint8)
# Add batch dimension to image and do inference on FP and INT8 models
input_image = np.expand_dims(image, 0)
res_fp = compiled_model_fp([input_image])
res_int8 = compiled_model_int8([input_image])
# Process inference outputs and convert to binary segementation masks
result_mask_fp = sigmoid(res_fp[output_layer_fp]).squeeze().round().astype(np.uint8)
result_mask_int8 = sigmoid(res_int8[output_layer_int8]).squeeze().round().astype(np.uint8)
result_mask_fp = rotate_and_flip(result_mask_fp)
result_mask_int8 = rotate_and_flip(result_mask_int8)
# Display images, annotations, FP32 result and INT8 result
ax[i, 0].imshow(display_image, cmap=colormap)
ax[i, 1].imshow(target_mask, cmap=colormap)
ax[i, 2].imshow(result_mask_fp, cmap=colormap)
ax[i, 3].imshow(result_mask_int8, cmap=colormap)
ax[i, 2].set_title("Prediction on FP32 model")
ax[i, 3].set_title("Prediction on INT8 model")
Visualizing results with seed 1707515536
Show Live Inference¶
To show live inference on the model in the notebook, we will use the asynchronous processing feature of OpenVINO.
We use the show_live_inference function from Notebook
Utils to show live inference. This
function uses Open Model
Zoo’s Async
Pipeline and Model API to perform asynchronous inference. After
inference on the specified CT scan has completed, the total time and
throughput (fps), including preprocessing and displaying, will be
printed.
NOTE: If you experience flickering on Firefox, consider using Chrome or Edge to run this notebook.
We load the segmentation model to OpenVINO Runtime with
SegmentationModel, based on the Open Model
Zoo Model API.
This model implementation includes pre and post processing for the
model. For SegmentationModel, this includes the code to create an
overlay of the segmentation mask on the original image/frame.
CASE = 117
segmentation_model = SegmentationModel(
ie=core, model_path=int8_ir_path, sigmoid=True, rotate_and_flip=True
)
case_path = BASEDIR / f"case_{CASE:05d}"
image_paths = sorted(case_path.glob("imaging_frames/*jpg"))
print(f"{case_path.name}, {len(image_paths)} images")
case_00117, 69 images
In the next cell, we run the show_live_inference function, which
loads the segmentation_model to the specified device (using
caching for faster model loading on GPU devices), loads the images,
performs inference, and displays the results on the frames loaded in
images in real-time.
# Possible options for device include "CPU", "GPU", "AUTO", "MULTI:CPU,GPU"
device = "CPU"
reader = LoadImage(image_only=True, dtype=np.uint8)
show_live_inference(
ie=core, image_paths=image_paths, model=segmentation_model, device=device, reader=reader
)
Loaded model to CPU in 0.18 seconds.
Total time for 68 frames: 2.68 seconds, fps:25.70
References¶
OpenVINO - NNCF Repository - Neural Network Compression Framework for fast model inference - OpenVINO API Tutorial - OpenVINO PyPI (pip install openvino-dev)
Kits19 Data - Kits19 Challenge Homepage - Kits19 GitHub Repository - The KiTS19 Challenge Data: 300 Kidney Tumor Cases with Clinical Context, CT Semantic Segmentations, and Surgical Outcomes - The state of the art in kidney and kidney tumor segmentation in contrast-enhanced CT imaging: Results of the KiTS19 challenge