Universal Segmentation with OneFormer and OpenVINO#
This Jupyter notebook can be launched after a local installation only.
This tutorial demonstrates how to use the OneFormer model from HuggingFace with OpenVINO. It describes how to download weights and create PyTorch model using Hugging Face transformers library, then convert model to OpenVINO Intermediate Representation format (IR) using OpenVINO Model Optimizer API and run model inference. Additionally, NNCF quantization is applied to improve OneFormer segmentation speed.
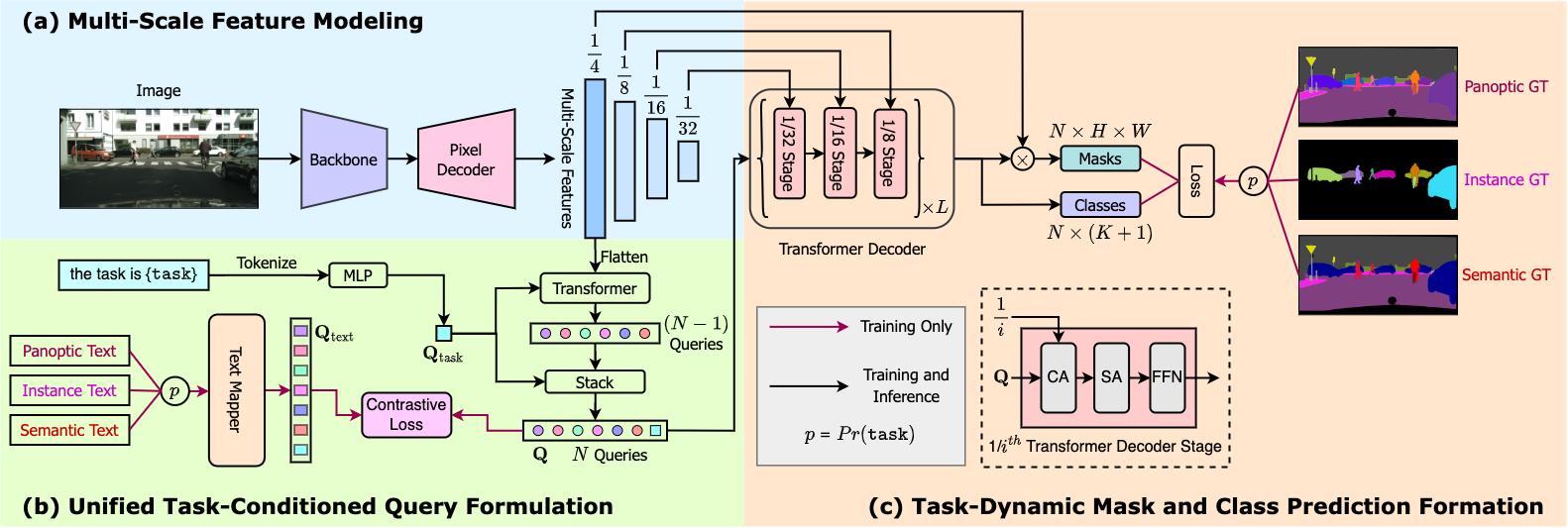
OneFormer is a follow-up work of Mask2Former. The latter still requires training on instance/semantic/panoptic datasets separately to get state-of-the-art results.
OneFormer incorporates a text module in the Mask2Former framework, to condition the model on the respective subtask (instance, semantic or panoptic). This gives even more accurate results, but comes with a cost of increased latency, however.
Table of contents:
Installation Instructions#
This is a self-contained example that relies solely on its own code.
We recommend running the notebook in a virtual environment. You only need a Jupyter server to start. For details, please refer to Installation Guide.
Install required libraries#
%pip install -q --extra-index-url https://download.pytorch.org/whl/cpu "transformers>=4.26.0" "openvino>=2023.1.0" "nncf>=2.7.0" "gradio>=4.19" "torch>=2.1" "matplotlib>=3.4" scipy ipywidgets Pillow tqdm
Prepare the environment#
Import all required packages and set paths for models and constant variables.
import warnings
from collections import defaultdict
from pathlib import Path
from transformers import OneFormerProcessor, OneFormerForUniversalSegmentation
from transformers.models.oneformer.modeling_oneformer import (
OneFormerForUniversalSegmentationOutput,
)
import torch
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
from PIL import Image
from PIL import ImageOps
import openvino
# Fetch `notebook_utils` module
import requests
if not Path("notebook_utils.py").exists():
r = requests.get(
url="https://raw.githubusercontent.com/openvinotoolkit/openvino_notebooks/latest/utils/notebook_utils.py",
)
open("notebook_utils.py", "w").write(r.text)
from notebook_utils import download_file, device_widget
# Read more about telemetry collection at https://github.com/openvinotoolkit/openvino_notebooks?tab=readme-ov-file#-telemetry
from notebook_utils import collect_telemetry
collect_telemetry("oneformer-segmentation.ipynb")
IR_PATH = Path("oneformer.xml")
OUTPUT_NAMES = ["class_queries_logits", "masks_queries_logits"]
Load OneFormer fine-tuned on COCO for universal segmentation#
Here we use the from_pretrained method of
OneFormerForUniversalSegmentation to load the HuggingFace OneFormer
model
based on Swin-L backbone and trained on
COCO dataset.
Also, we use HuggingFace processor to prepare the model inputs from images and post-process model outputs for visualization.
processor = OneFormerProcessor.from_pretrained("shi-labs/oneformer_coco_swin_large")
model = OneFormerForUniversalSegmentation.from_pretrained(
"shi-labs/oneformer_coco_swin_large",
)
id2label = model.config.id2label
task_seq_length = processor.task_seq_length
shape = (800, 800)
dummy_input = {
"pixel_values": torch.randn(1, 3, *shape),
"task_inputs": torch.randn(1, task_seq_length),
}
Convert the model to OpenVINO IR format#
Convert the PyTorch model to IR format to take advantage of OpenVINO
optimization tools and features. The openvino.convert_model python
function in OpenVINO Converter can convert the model. The function
returns instance of OpenVINO Model class, which is ready to use in
Python interface. However, it can also be serialized to OpenVINO IR
format for future execution using save_model function. PyTorch to
OpenVINO conversion is based on TorchScript tracing. HuggingFace models
have specific configuration parameter torchscript, which can be used
for making the model more suitable for tracing. For preparing model. we
should provide PyTorch model instance and example input to
openvino.convert_model.
model.config.torchscript = True
if not IR_PATH.exists():
with warnings.catch_warnings():
warnings.simplefilter("ignore")
model = openvino.convert_model(model, example_input=dummy_input)
openvino.save_model(model, IR_PATH, compress_to_fp16=False)
Select inference device#
Select device from dropdown list for running inference using OpenVINO
core = openvino.Core()
device = device_widget()
device
Dropdown(description='Device:', index=1, options=('CPU', 'AUTO'), value='AUTO')
We can prepare the image using the HuggingFace processor. OneFormer leverages a processor which internally consists of an image processor (for the image modality) and a tokenizer (for the text modality). OneFormer is actually a multimodal model, since it incorporates both images and text to solve image segmentation.
def prepare_inputs(image: Image.Image, task: str):
"""Convert image to model input"""
image = ImageOps.pad(image, shape)
inputs = processor(image, [task], return_tensors="pt")
converted = {
"pixel_values": inputs["pixel_values"],
"task_inputs": inputs["task_inputs"],
}
return converted
def process_output(d):
"""Convert OpenVINO model output to HuggingFace representation for visualization"""
hf_kwargs = {output_name: torch.tensor(d[output_name]) for output_name in OUTPUT_NAMES}
return OneFormerForUniversalSegmentationOutput(**hf_kwargs)
# Read the model from files.
model = core.read_model(model=IR_PATH)
# Compile the model.
compiled_model = core.compile_model(model=model, device_name=device.value)
Model predicts class_queries_logits of shape
(batch_size, num_queries) and masks_queries_logits of shape
(batch_size, num_queries, height, width).
Here we define functions for visualization of network outputs to show the inference results.
class Visualizer:
@staticmethod
def extract_legend(handles):
fig = plt.figure()
fig.legend(handles=handles, ncol=len(handles) // 20 + 1, loc="center")
fig.tight_layout()
return fig
@staticmethod
def predicted_semantic_map_to_figure(predicted_map):
segmentation = predicted_map[0]
# get the used color map
viridis = plt.get_cmap("viridis", max(1, torch.max(segmentation)))
# get all the unique numbers
labels_ids = torch.unique(segmentation).tolist()
fig, ax = plt.subplots()
ax.imshow(segmentation)
ax.set_axis_off()
handles = []
for label_id in labels_ids:
label = id2label[label_id]
color = viridis(label_id)
handles.append(mpatches.Patch(color=color, label=label))
fig_legend = Visualizer.extract_legend(handles=handles)
fig.tight_layout()
return fig, fig_legend
@staticmethod
def predicted_instance_map_to_figure(predicted_map):
segmentation = predicted_map[0]["segmentation"]
segments_info = predicted_map[0]["segments_info"]
# get the used color map
viridis = plt.get_cmap("viridis", max(torch.max(segmentation), 1))
fig, ax = plt.subplots()
ax.imshow(segmentation)
ax.set_axis_off()
instances_counter = defaultdict(int)
handles = []
# for each segment, draw its legend
for segment in segments_info:
segment_id = segment["id"]
segment_label_id = segment["label_id"]
segment_label = id2label[segment_label_id]
label = f"{segment_label}-{instances_counter[segment_label_id]}"
instances_counter[segment_label_id] += 1
color = viridis(segment_id)
handles.append(mpatches.Patch(color=color, label=label))
fig_legend = Visualizer.extract_legend(handles)
fig.tight_layout()
return fig, fig_legend
@staticmethod
def predicted_panoptic_map_to_figure(predicted_map):
segmentation = predicted_map[0]["segmentation"]
segments_info = predicted_map[0]["segments_info"]
# get the used color map
viridis = plt.get_cmap("viridis", max(torch.max(segmentation), 1))
fig, ax = plt.subplots()
ax.imshow(segmentation)
ax.set_axis_off()
instances_counter = defaultdict(int)
handles = []
# for each segment, draw its legend
for segment in segments_info:
segment_id = segment["id"]
segment_label_id = segment["label_id"]
segment_label = id2label[segment_label_id]
label = f"{segment_label}-{instances_counter[segment_label_id]}"
instances_counter[segment_label_id] += 1
color = viridis(segment_id)
handles.append(mpatches.Patch(color=color, label=label))
fig_legend = Visualizer.extract_legend(handles)
fig.tight_layout()
return fig, fig_legend
@staticmethod
def figures_to_images(fig, fig_legend, name_suffix=""):
seg_filename, leg_filename = (
f"segmentation{name_suffix}.png",
f"legend{name_suffix}.png",
)
fig.savefig(seg_filename, bbox_inches="tight")
fig_legend.savefig(leg_filename, bbox_inches="tight")
segmentation = Image.open(seg_filename)
legend = Image.open(leg_filename)
return segmentation, legend
def segment(model, img: Image.Image, task: str):
"""
Apply segmentation on an image.
Args:
img: Input image. It will be resized to 800x800.
task: String describing the segmentation task. Supported values are: "semantic", "instance" and "panoptic".
Returns:
Tuple[Figure, Figure]: Segmentation map and legend charts.
"""
if img is None:
raise gr.Error("Please load the image or use one from the examples list")
inputs = prepare_inputs(img, task)
outputs = model(inputs)
hf_output = process_output(outputs)
predicted_map = getattr(processor, f"post_process_{task}_segmentation")(hf_output, target_sizes=[img.size[::-1]])
return getattr(Visualizer, f"predicted_{task}_map_to_figure")(predicted_map)
if not Path("sample.jpg").exists():
download_file("http://images.cocodataset.org/val2017/000000439180.jpg", "sample.jpg")
image = Image.open("sample.jpg")
image
sample.jpg: 0%| | 0.00/194k [00:00<?, ?B/s]

Choose a segmentation task#
from ipywidgets import Dropdown
task = Dropdown(options=["semantic", "instance", "panoptic"], value="semantic")
task
Dropdown(options=('semantic', 'instance', 'panoptic'), value='semantic')
Inference#
import matplotlib
matplotlib.use("Agg") # disable showing figures
def stack_images_horizontally(img1: Image, img2: Image):
res = Image.new("RGB", (img1.width + img2.width, max(img1.height, img2.height)), (255, 255, 255))
res.paste(img1, (0, 0))
res.paste(img2, (img1.width, 0))
return res
segmentation_fig, legend_fig = segment(compiled_model, image, task.value)
segmentation_image, legend_image = Visualizer.figures_to_images(segmentation_fig, legend_fig)
plt.close("all")
prediction = stack_images_horizontally(segmentation_image, legend_image)
prediction

Quantization#
NNCF enables
post-training quantization by adding quantization layers into model
graph and then using a subset of the training dataset to initialize the
parameters of these additional quantization layers. Quantized operations
are executed in INT8 instead of FP32/FP16 making model
inference faster.
The optimization process contains the following steps: 1. Create a
calibration dataset for quantization. 2. Run nncf.quantize() to
obtain quantized model. 3. Serialize the INT8 model using
openvino.save_model() function.
Note: Quantization is time and memory consuming operation. Running quantization code below may take some time.
Please select below whether you would like to run quantization to improve model inference speed.
from notebook_utils import quantization_widget
compiled_quantized_model = None
to_quantize = quantization_widget(False)
to_quantize
Checkbox(value=True, description='Quantization')
Let’s load skip magic extension to skip quantization if to_quantize is not selected
# Fetch `skip_kernel_extension` module
if not Path("skip_kernel_extension.py").exists():
r = requests.get(
url="https://raw.githubusercontent.com/openvinotoolkit/openvino_notebooks/latest/utils/skip_kernel_extension.py",
)
open("skip_kernel_extension.py", "w").write(r.text)
%load_ext skip_kernel_extension
Preparing calibration dataset#
We use images from COCO128 dataset as calibration samples.
%%skip not $to_quantize.value
import nncf
import torch.utils.data as data
from zipfile import ZipFile
DATA_URL = "https://ultralytics.com/assets/coco128.zip"
OUT_DIR = Path('.')
class COCOLoader(data.Dataset):
def __init__(self, images_path):
self.images = list(Path(images_path).iterdir())
def __getitem__(self, index):
image = Image.open(self.images[index])
if image.mode == 'L':
rgb_image = Image.new("RGB", image.size)
rgb_image.paste(image)
image = rgb_image
return image
def __len__(self):
return len(self.images)
def download_coco128_dataset():
download_file(DATA_URL, directory=OUT_DIR, show_progress=True)
if not (OUT_DIR / "coco128/images/train2017").exists():
with ZipFile('coco128.zip' , "r") as zip_ref:
zip_ref.extractall(OUT_DIR)
coco_dataset = COCOLoader(OUT_DIR / 'coco128/images/train2017')
return coco_dataset
def transform_fn(image):
# We quantize model in panoptic mode because it produces optimal results for both semantic and instance segmentation tasks
inputs = prepare_inputs(image, "panoptic")
return inputs
coco_dataset = download_coco128_dataset()
calibration_dataset = nncf.Dataset(coco_dataset, transform_fn)
INFO:nncf:NNCF initialized successfully. Supported frameworks detected: torch, tensorflow, onnx, openvino
coco128.zip: 0%| | 0.00/6.66M [00:00<?, ?B/s]
Run quantization#
Below we call nncf.quantize() in order to apply quantization to
OneFormer model.
%%skip not $to_quantize.value
INT8_IR_PATH = Path(str(IR_PATH).replace(".xml", "_int8.xml"))
if not INT8_IR_PATH.exists():
quantized_model = nncf.quantize(
model,
calibration_dataset,
model_type=nncf.parameters.ModelType.TRANSFORMER,
subset_size=len(coco_dataset),
# smooth_quant_alpha value of 0.5 was selected based on prediction quality visual examination
advanced_parameters=nncf.AdvancedQuantizationParameters(smooth_quant_alpha=0.5))
openvino.save_model(quantized_model, INT8_IR_PATH)
else:
quantized_model = core.read_model(INT8_IR_PATH)
compiled_quantized_model = core.compile_model(model=quantized_model, device_name=device.value)
Statistics collection: 100%|██████████████████████████████████████████████████████████████████████████████████████████████| 128/128 [03:55<00:00, 1.84s/it]
Applying Smooth Quant: 100%|██████████████████████████████████████████████████████████████████████████████████████████████| 216/216 [00:18<00:00, 11.89it/s]
INFO:nncf:105 ignored nodes was found by name in the NNCFGraph
Statistics collection: 100%|██████████████████████████████████████████████████████████████████████████████████████████████| 128/128 [09:24<00:00, 4.41s/it]
Applying Fast Bias correction: 100%|██████████████████████████████████████████████████████████████████████████████████████| 338/338 [03:20<00:00, 1.68it/s]
Let’s see quantized model prediction next to original model prediction.
%%skip not $to_quantize.value
from IPython.display import display
image = Image.open("sample.jpg")
segmentation_fig, legend_fig = segment(compiled_quantized_model, image, task.value)
segmentation_image, legend_image = Visualizer.figures_to_images(segmentation_fig, legend_fig, name_suffix="_int8")
plt.close("all")
prediction_int8 = stack_images_horizontally(segmentation_image, legend_image)
print("Original model prediction:")
display(prediction)
print("Quantized model prediction:")
display(prediction_int8)
Original model prediction:

Quantized model prediction:

Compare model size and performance#
Below we compare original and quantized model footprint and inference speed.
%%skip not $to_quantize.value
import time
import numpy as np
from tqdm.auto import tqdm
INFERENCE_TIME_DATASET_SIZE = 30
def calculate_compression_rate(model_path_ov, model_path_ov_int8):
model_size_fp32 = model_path_ov.with_suffix(".bin").stat().st_size / 1024
model_size_int8 = model_path_ov_int8.with_suffix(".bin").stat().st_size / 1024
print("Model footprint comparison:")
print(f" * FP32 IR model size: {model_size_fp32:.2f} KB")
print(f" * INT8 IR model size: {model_size_int8:.2f} KB")
return model_size_fp32, model_size_int8
def calculate_call_inference_time(model):
inference_time = []
for i in tqdm(range(INFERENCE_TIME_DATASET_SIZE), desc="Measuring performance"):
image = coco_dataset[i]
start = time.perf_counter()
segment(model, image, task.value)
end = time.perf_counter()
delta = end - start
inference_time.append(delta)
return np.median(inference_time)
time_fp32 = calculate_call_inference_time(compiled_model)
time_int8 = calculate_call_inference_time(compiled_quantized_model)
model_size_fp32, model_size_int8 = calculate_compression_rate(IR_PATH, INT8_IR_PATH)
print(f"Model footprint reduction: {model_size_fp32 / model_size_int8:.3f}")
print(f"Performance speedup: {time_fp32 / time_int8:.3f}")
Measuring performance: 0%| | 0/30 [00:00<?, ?it/s]
Measuring performance: 0%| | 0/30 [00:00<?, ?it/s]
Model footprint comparison:
* FP32 IR model size: 899385.45 KB
* INT8 IR model size: 237545.83 KB
Model footprint reduction: 3.786
Performance speedup: 1.260
Interactive Demo#
import time
quantized_model_present = compiled_quantized_model is not None
def compile_model(device):
global compiled_model
global compiled_quantized_model
compiled_model = core.compile_model(model=model, device_name=device)
if quantized_model_present:
compiled_quantized_model = core.compile_model(model=quantized_model, device_name=device)
def segment_wrapper(image, task, run_quantized=False):
current_model = compiled_quantized_model if run_quantized else compiled_model
start_time = time.perf_counter()
segmentation_fig, legend_fig = segment(current_model, image, task)
end_time = time.perf_counter()
name_suffix = "" if not quantized_model_present else "_int8" if run_quantized else "_fp32"
segmentation_image, legend_image = Visualizer.figures_to_images(segmentation_fig, legend_fig, name_suffix=name_suffix)
plt.close("all")
result = stack_images_horizontally(segmentation_image, legend_image)
return result, f"{end_time - start_time:.2f}"
if not Path("gradio_helper.py").exists():
r = requests.get(url="https://raw.githubusercontent.com/openvinotoolkit/openvino_notebooks/latest/notebooks/oneformer-segmentation/gradio_helper.py")
open("gradio_helper.py", "w").write(r.text)
from gradio_helper import make_demo
demo = make_demo(run_fn=segment_wrapper, compile_model_fn=compile_model, quantized=quantized_model_present)
try:
demo.launch(debug=False)
except Exception:
demo.launch(share=True, debug=False)
# if you are launching remotely, specify server_name and server_port
# demo.launch(server_name='your server name', server_port='server port in int')
# Read more in the docs: https://gradio.app/docs/